Wright Lab
Neurogenomics Research
Hello there, welcome to the Wright Lab! We are a human genetics research group at the University of Manitoba that uses genomics to understand the brain in health and disease.
We would like to understand why individuals affected by the same neurological disorder can experience differences in disease severity and onset. Insights obtained from these analyses can be used to guide future therapeutic approaches to improve disease management.

About
Genetic variants that alter the clinical expression of disease-causing mutations are known as genetic modifiers and can help explain differences in disease outcomes. For example, genetic variants in genes from DNA repair pathways influence age-of-onset for several neurological diseases.
We use large-scale bioinformatic analyses to inform functional genomic experiments. This includes using gene editing and human stem-cell-derived neural models to study genetic modifiers of neurological disorders.
Such findings can lead to a better understanding of human biology, the identification of therapeutic targets, and improved risk prediction models.
KEY PROJECTS
- Genetic modifiers of neurological disorders
- Genomics of DNA repair in the brain
Territory acknowledgement: Our research group is located on the original lands of Anishinaabeg, Cree, Oji-Cree, Dakota and Dene peoples, and on the homeland of the Métis Nation. We respect the Treaties that were made on these territories, we acknowledge the harms and mistakes of the past, and we dedicate ourselves to moving forward in partnership with Indigenous communities in a spirit of reconciliation and collaboration.
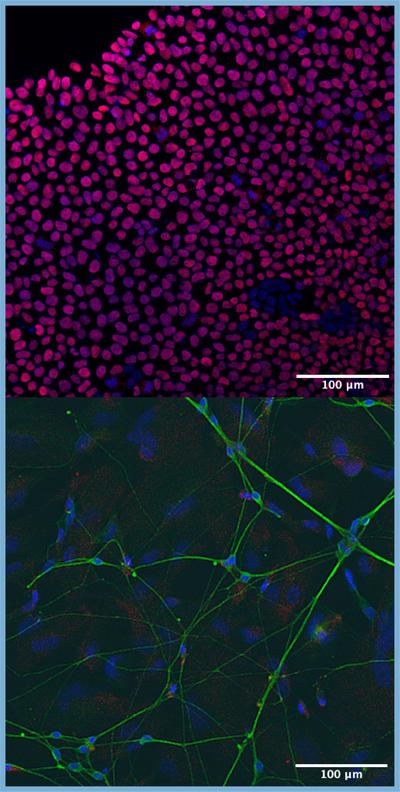
What we do
We use both wet and dry lab approaches to understand human biology
• Huntington disease
• CANVAS
• Rett syndrome
• Genomic fine mapping
• Single-cell sequencing
• Machine learning
• Human stem cells
• Neural cell culture
• CRISPR gene editing
Affiliations
We are primarily affiliated with the Department of Pharmacology & Therapeutics at the University of Manitoba. Our lab is in the PrairieNeuro Research Centre at the Kleysen Institute for Advanced Medicine, Health Sciences Centre. We are also affiliated with the Department of Biochemistry and Medical Genetics, as well as the Children’s Hospital Research Institute of Manitoba (CHRIM, ENRRICH theme).

Latest News

We are excited that the first pieces of CFI equipment for our Precision Genomics Suite have started to arrive! This includes the Chromium iX, Visium CytAssist and Xenium Analyzer. We just need to complete a couple of renovations to the room that will house the equipment and look forward to receiving training in the new year.

Mason was funded this Summer through the new studentship offered through the Excellence in Neurodevelopment and Rehabilitation Research in Child Health (ENRRICH) Theme at CHRIM. They recently interviewed her about her experiences over the last few months and posted it on their blog.

Prats received the Stem Cell Network Summer Studentship Award to support his work in the lab over the Summer. Notably, he was the only Manitoban student to receive this award!

We are excited to announce that we have received new infrastructure funding from the Canada Foundation for Innovation (CFI) John R. Evans Leaders Fund (JELF)! This funding will be used to develop a Precision Genomics Suite at the university that will allow us to perform extensive genomic studies at the resolution of individual cells.
Tweets by GalenWright






